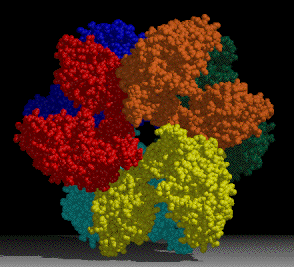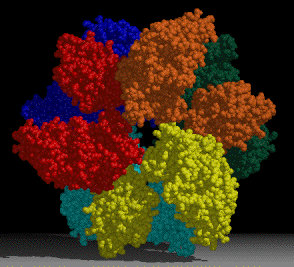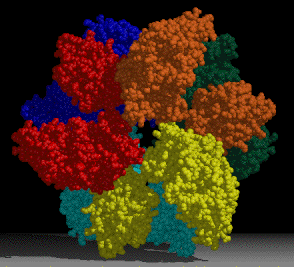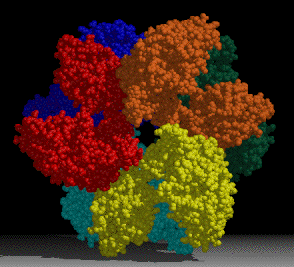
A project of interest in the lab is understanding sulfur assimilation, or fixation. This project focuses on the structural features underlying the catalytic and regulatory properties of the sulfate activating enzymes, ATP sulfurylase, APS kinase, and PAPS Reductase, which catalyze in order, the reactions:

where: APS = adenosine 5′-phosphosulfate (adenylylsulfate); PAPS = 3′-phosphoadenosine-5′-phosphosulfate (3′-phosphoadenylylsulfate); PAP = adenosine 3′,5′-bisphosphate, and TRX is Thioredoxin.
The first two enzymes in the pathway (ATP Sulfurylase & APS Kinase) are conserved in all organisms to activate sulfate into the universal sulfate donor (PAPS) used to sulfurylate many diverse biomolecules including proteins, lipids, and carbohydrates. The last step in sulfur assimilation, PAPS reductase, is not conserved in many higher eukaryotes, such as humans. Our lab is exploring the structural basis for sulfate activation and sulfur assimilation in many organisms including bacterial, archaea, fungi, and humans, to better understand how each organism as adapted the enzymes to suit the physiological function of the pathway. Some chemolithotrophic organisms use the pathway in the final oxidation of sulfur to harness energy. Additionally, we are investigating these enzymes in Mycobacterium tuberculosis, because many of the enzymes in the pathway display sequence and structural features different than the orthologous human enzymes, making them potential therapeutic targets to treat tuberculosis.
