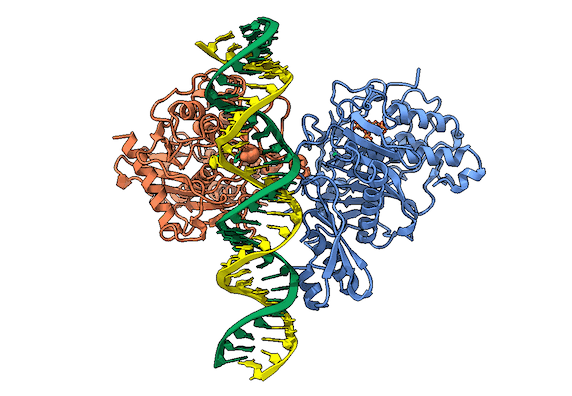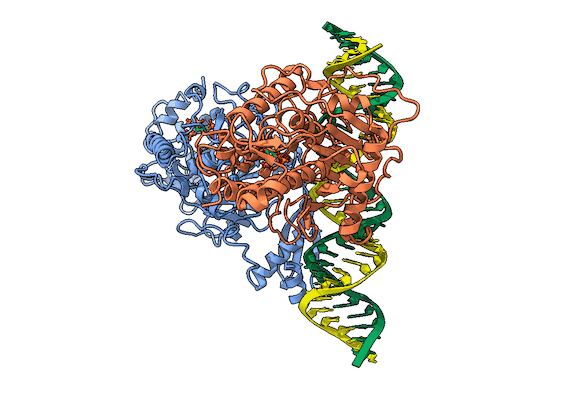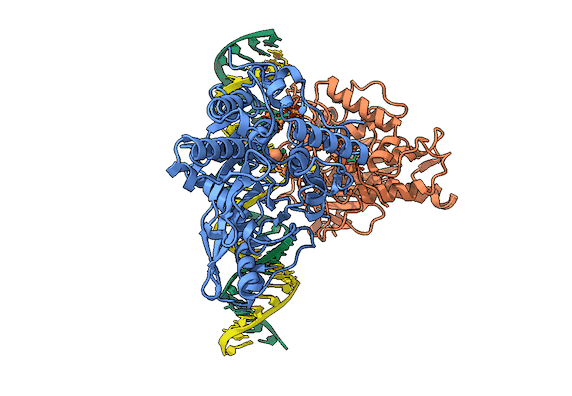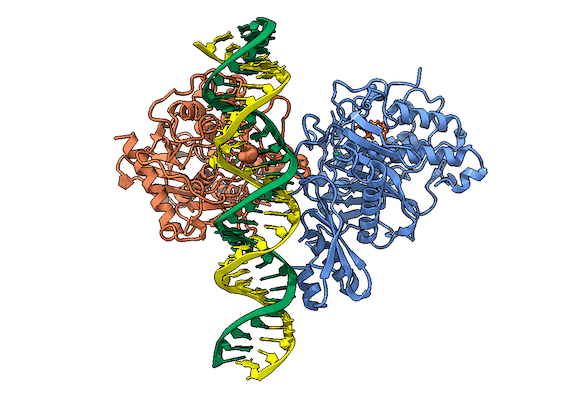
RNA is frequently edited after being transcribed by RNA polymerase. One of the most abundant RNA modifications found in metazoans is the deamination of adenosine to inosine (A-to-I). This reaction is catalyzed by the class of enzymes called adenosine deaminases acting on dsRNA (ADAR).
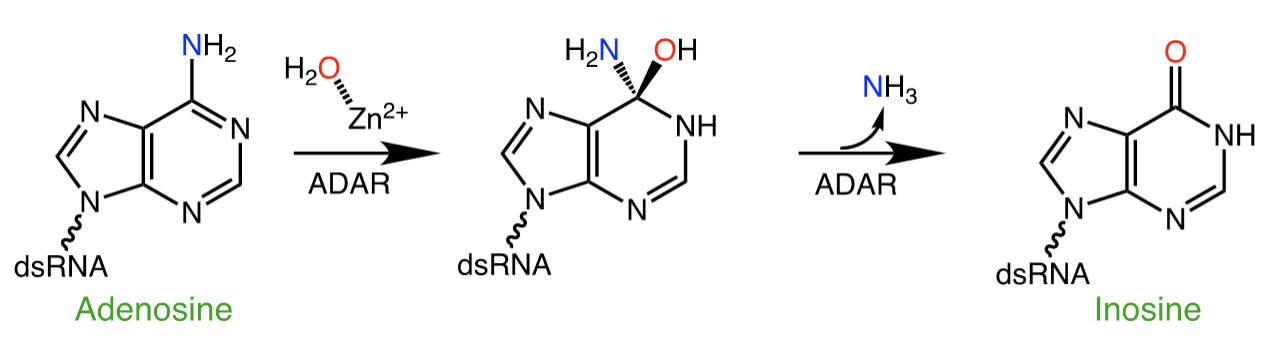
In the ADAR deamination reaction, the resulting inosine nucleotide product behaves more like guanosine in hydrogen-bonding patterns, consequently A-to-I edits can significantly alter the RNA function including; recoding mRNA, altering splice sites, and changing RNA secondary structure. Over three million RNA A-to-I edits have been documented in healthy human tissues. RNA editing is critical for normal cellular function and dysregulated ADAR activity can resulted in a variety of neurological disorders such as epilepsy and Prader Willi Syndrome, depression, schizophrenia, amyotrophic lateral sclerosis, as well as cancer
In collaboration with the Beal lab, we have structurally and biochemically characterized human ADAR2 deaminase domain bound to a 23mer RNA duplex, which contained the nucleoside analog 8-azanebularine in place of adenine at the edited site. Upon bonding in the ADAR active site, 8-azanebularine becomes hydrated mimicking the proposed intermediate thus forming a tight-binding dsRNA-ADAR complex amenable for structural studies.
We are currently continuing our studies on investigating the structural basis for RNA selectivity and mechanism by looking at different ADARs and ADAR constructs bound to RNA.
